Customising map appearance
Source:vignettes/customising-map-appearance.Rmd
customising-map-appearance.RmdIn this article we will describe different ways to customise the appearance of an antigenic map with for example different point colours, shapes and sizes. We’ll first describe the key functions and then go through an example.
Main point style functions
The main functions for controlling point style parameters are:
-
agFill()andsrFill()for controlling the antigen and sera fill colour -
agOutline()andsrOutline()for controlling the antigen and sera outline colour -
agOutlineWidth()andsrOutlineWidth()for controlling the antigen and sera outline stroke width -
agSize()andsrSize()for controlling the antigen and sera point sizes -
agShape()andsrShape()for controlling the antigen and sera point shapes -
agShown()andsrShown()for controlling whether antigen and serum points are visible
Note that the primary supported shapes values are “circle”, “box” and “triangle”, but see below for details on further map customisation.
There is also the function ptDrawingOrder() which
controls the order in which points are plotted according to their index
in the map (numbering the antigens first then the sera).
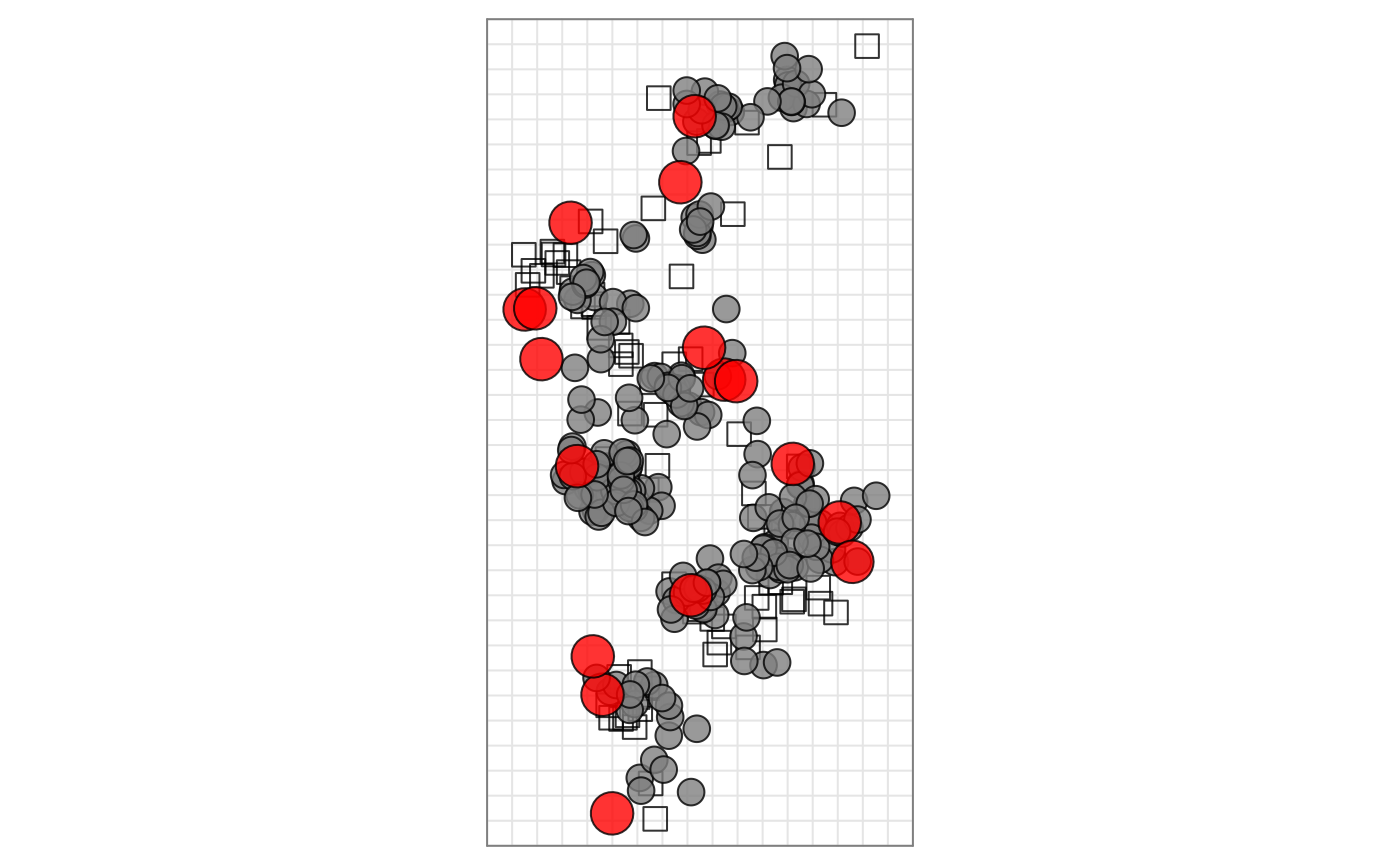
An example of styling a map
library(Racmacs)
map <- read.acmap(system.file("extdata/h3map2004.ace", package = "Racmacs"))
# Rotate the map
map <- rotateMap(map, 210)
# Colour all the antigens grey
agFill(map) <- "grey50"
# Colour all the vaccine antigens in red and increase their size
vaccine_strains <- c(
"PC/1/73",
"VI/3/75",
"TE/1/77",
"BA/1/79",
"PH/2/82",
"LE/360/86",
"SH/11/87",
"SI/2/87",
"BE/353/89",
"GU/54/89",
"BE/32/92",
"SD/9/93",
"JO/33/94",
"WU/359/95",
"SY/5/97",
"MW/10/99",
"FU/411/02"
)
vaccine_ags <- agNames(map) %in% vaccine_strains
agFill(map)[vaccine_ags] <- "red"
agSize(map)[vaccine_ags] <- 8
# Bring the antigens to the top of the plot
ptDrawingOrder(map) <- c(
seq_len(numSera(map)) + numAntigens(map), # Draw sera first
which(!vaccine_ags), # Then draw non vaccine ags
which(vaccine_ags) # The draw vaccine ags
)
# Plot the map
plot(map)